Hyperuniformity
Functions designed to study the hyperuniformity of a stationary point process using estimation(s) of its structure factor StructureFactor.
Hyperuniformity diagnostics:
multiscale_test(): Statistical test of hyperuniformity asymptomatically valid.
effective_hyperuniformity(): Test of effective hyperuniformity.
hyperuniformity_class(): Estimation of the possible class of hyperuniformity.
- Additional functions:
bin_data(): Method for regularizing structure factor’s estimation.subwindows(): Method for generating a list of subwindows from a father window with the corresponding minimum allowed wavevectors (or wavenumbers).
Note
Typical usage
Test the hyperuniformity using the statistical test
multiscale_test()or the test of effective hyperuniformityeffective_hyperuniformity().If the hyperuniformity was approved, find the possible hyperuniformity class using
hyperuniformity_class().
- structure_factor.hyperuniformity.multiscale_test(point_pattern_list, estimator, subwindows_list, k_list, mean_poisson, m_list=None, proba_list=None, verbose=False, **kwargs)[source]
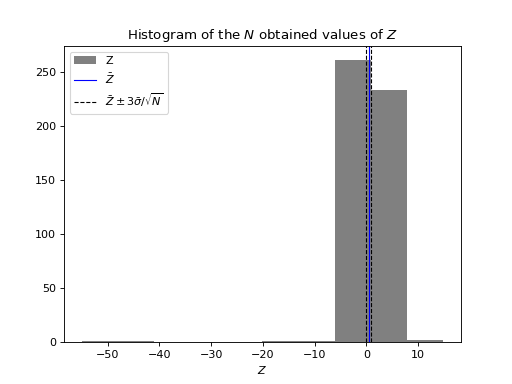
Compute the sample mean \(\bar{Z}\) and the corresponding standard deviation \(\bar{\sigma}/\sqrt{N}\) of the coupled sum estimator \(Z\) of a point process using a list of \(N\) PointPatterns and \(N\) realizations from the random variable \(M\).
The test rejects the hyperuniformity hypothesis if the confidence interval \(CI[\mathbb{E}[Z]]= [\bar{Z} - z \bar{\sigma}/\sqrt{N}, \bar{Z} + z \bar{\sigma}/\sqrt{N}]\), doesn’t contain zero, and vice-versa. (See the multiscale hyperuniformity test in [HGBLachiezeR22]).
- Parameters
point_pattern_list (list) – List of
PointPattern(s). Each element of the list is an encapsulation of a realization of a point process, the observation window, and (optionally) the intensity of the point process. AllPointPattern(s) should have the same window and intensity.estimator (str) – Choice of structure factor’s estimator. The parameters of the chosen estimator must be added as keyword arguments. The available estimators are “scattering_intensity”, “tapered_estimator”, “bartlett_isotropic_estimator”, and “quadrature_estimator_isotropic”. See
StructureFactor.subwindows_list (list) – List of increasing cubic or ball-shaped
AbstractSpatialWindow, typically, obtained usingsubwindows(). The shape of the windows depends on the choice of theestimator. Each element ofpoint_pattern_listwill be restricted to these windows to compute \(Z\).k_list (list) – List of wavevectors (or wavenumbers) where the
estimatoris to be evaluated. Each element is associated with an element ofsubwindows_list. Typically, obtained usingsubwindows().mean_poisson (int) – Parameter of the Poisson r.v. \(M\) used to compute \(Z\). To use a different distribution of the r.v. \(M\), set
mean_poisson=Noneand specifym_listandproba_listcorresponding to \(M\).m_list (list, optional) – List of positive integer realizations of the r.v. \(M\), used when
mean_poisson=None. Each element ofm_listis associated with an element ofpoint_pattern_listto compute a realization of \(Z\). Defaults to None.proba_list (list, optional) – List of \(\mathbb{P}(M \geq j)\) used with
m_listwhenmean_poisson=None. Should contains at leastmax(m_list)elements. Defaults to None.verbose (bool, optional) – If “True” and
mean_poissonis not None, print the re-sampled values of \(M\). Defaults to False.
- Keyword Arguments
kwargs (dict) – Parameters of the chosen
estimatorof the structure factor. SeeStructureFactor.- Returns
“mean_Z”: The sample mean of \(Z\).
”std_mean_Z”: The sample standard deviation of \(Z\) divided by the square root of the number of samples.
”Z”: The obtained values of \(Z\).
”M”: The used values of \(M\).
- Return type
dict(float, float, list, list)
Example
import numpy as np from structure_factor.point_processes import HomogeneousPoissonPointProcess from structure_factor.spatial_windows import BoxWindow from structure_factor.hyperuniformity import subwindows, multiscale_test # PointPattern list poisson = HomogeneousPoissonPointProcess(intensity=1 / np.pi) N = 500 L = 160 window = BoxWindow([[-L / 2, L / 2]] * 2) point_patterns = [ poisson.generate_point_pattern(window=window) for _ in range(N) ] # subwindows and k l_0 = 40 subwindows_list, k = subwindows(window, subwindows_type="BoxWindow", param_0=l_0) # multiscale hyperuniformity test mean_poisson = 90 summary = multiscale_test(point_patterns, estimator="scattering_intensity", k_list=k, subwindows_list=subwindows_list, mean_poisson=mean_poisson, verbose=True) import matplotlib.pyplot as plt plt.hist(summary["Z"], color="grey", label="Z") plt.axvline(summary["mean_Z"], color="b", linewidth=1, label=r"$\bar{Z}$") plt.axvline(summary["mean_Z"] - 3*summary["std_mean_Z"], linestyle='dashed', color="k", linewidth=1, label=r"$\bar{Z} \pm 3 \bar{\sigma}/ \sqrt{N}$") plt.axvline(summary["mean_Z"] + 3*summary["std_mean_Z"], linestyle='dashed', color="k", linewidth=1) plt.xlabel(r"$Z$") plt.title(r"Histogram of the $N$ obtained values of $Z$") plt.legend() plt.show()
(Source code, png, hires.png, pdf)
 Definition
DefinitionLet \(\mathcal{X} \in \mathbb{R}^d\) be a stationary point process of which we consider an increasing sequence of sets \((\mathcal{X} \cap W_m)_{m \geq 1}\), with \((W_m)_m\) centered box (or ball)-shaped windows s.t. \(W_s \subset W_r\) for all \(0< s<r\), and \(W_{\infty} = \mathbb{R}^d\). We define the sequence of r.v. \(Y_m = 1\wedge \widehat{S}_m(\mathbf{k}_m^{\text{min}})\), where \(\widehat{S}_m\) is one of the positive, asymptotically unbiased estimators of the structure factor of \(\mathcal{X}\) applied on the observation \(\mathcal{X} \cap W_m\), and \(\mathbf{k}_m^{\text{min}}\) is the minimum allowed wavevector associated with \(W_m\).
Under some assumptions ([HGBLachiezeR22], Section 4) \(\mathcal{X}\) is hyperuniform iff \(\mathbb{E}[Z]=0\). Where \(Z\) is the coupled sum estimator of [RG15] defined by,
\[Z = \sum_{j=1}^{M} \frac{Y_j - Y_{j-1}}{\mathbb{P}(M\geq j)},\]with \(M\) an \(\mathbb{N}\)-valued random variable such that \(\mathbb{P}(M \geq j)>0\) for all \(j\), and \(Y_{0}=0\).
Important
If
mean_poissonis not None, there is a step of accepting/rejecting while sampling from the r.v. \(M\). If the biggest subwindow associated with \(M'\) (obtained value of \(M\)) is larger than the father window, then \(M'\) is rejected, and we resample from \(M\). Typically,mean_poissonshould be chosen s.t. the probability that the biggest subwindow is larger than the father window is small enough.The test is asymptotically valid, so it might fail in diagnosing hyperuniformity if the number of samples used to compute \(\bar{Z}\) or \(\lambda\) is too small.
Note
Typical usage
The function
subwindows()can be used to generate from a father window a list of subwindows and the associated allowed wavevectors/wavenumbers.
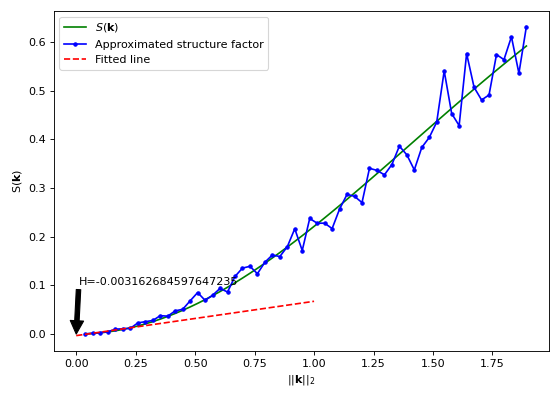
- structure_factor.hyperuniformity.effective_hyperuniformity(k_norm, sf, k_norm_stop, std_sf=None, **kwargs)[source]
Evaluate the index \(H\) of hyperuniformity of a point process using its structure factor
sf. If \(H<10^{-3}\) the corresponding point process is deemed effectively hyperuniform.- Parameters
k_norm (numpy.ndarray) – Vector of wavenumbers (i.e. norms of wavevectors).
sf (numpy.ndarray) – Evaluations of the structure factor, of the given point process, at
k_norm.std_sf (numpy.ndarray, optional) – Standard deviations associated with
sf. Defaults to None.k_norm_stop (float) – Threshold on
k_norm. Used to find the numerator of \(H\) by linear regression ofsfup to the value associated withk_norm_stop.
- Keyword Arguments
kwargs (dict) – Keyword arguments (except “sigma”) of scipy.scipy.optimize.curve_fit parameters.
- Returns
“H”: Value of the index \(H\).
”s0_std”: Standard deviation of the numerator of \(H\).
”fitted_line”: Line used to find the numerator of \(H\).
”idx_first_peak”: Index of \(k_{peak}\) if it exists.
- Return type
dict(float, float, function, int)
Example
import matplotlib.pyplot as plt import numpy as np from structure_factor.data import load_data from structure_factor.hyperuniformity import effective_hyperuniformity from structure_factor.point_processes import GinibrePointProcess from structure_factor.structure_factor import StructureFactor from structure_factor.tapered_estimators_isotropic import ( allowed_k_norm_bartlett_isotropic, ) point_pattern = load_data.load_ginibre() point_process = GinibrePointProcess() sf = StructureFactor(point_pattern) d, r = point_pattern.dimension, point_pattern.window.radius k_norm = allowed_k_norm_bartlett_isotropic(dimension=d, radius=r, nb_values=60) k_norm, sf_estimated = sf.bartlett_isotropic_estimator(k_norm) summary = effective_hyperuniformity(k_norm, sf_estimated, k_norm_stop=0.2) x = np.linspace(0, 1, 300) sf_fitted_line = summary["fitted_line"](x) sf_theoretical = point_process.structure_factor(k_norm) fig, ax = plt.subplots(figsize=(7, 5)) ax.plot(k_norm, sf_theoretical, "g", label=r"$S(\mathbf{k})$") ax.plot(k_norm, sf_estimated, "b", marker=".", label="Approximated structure factor") ax.plot(x, sf_fitted_line, "r--", label="Fitted line") ax.annotate( "H={}".format(summary["H"]), xy=(0, 0), xytext=(0.01, 0.1), arrowprops=dict(facecolor="black", shrink=0.0001), ) ax.legend() ax.set_xlabel(r"$||\mathbf{k}||_2$") ax.set_ylabel(r"$\mathsf{S}(\mathbf{k})$") plt.tight_layout(pad=1)
(Source code, png, hires.png, pdf)
 Definition
DefinitionA stationary isotropic point process \(\mathcal{X} \subset \mathbb{R}^d\), is said to be effectively hyperuniform if \(H \leq 10^{-3}\) where \(H\) is defined following [Tor18] (Section 11.1.6) and [KLovricC+19] (supplementary Section 8) by,
\[H = \frac{\hat{S}(\mathbf{0})}{S(\mathbf{k}_{peak})}\cdot\]\(S\) is the structure factor of \(\mathcal{X}\),
\(\hat{S}(\mathbf{0})\) is a linear extrapolation of the structure factor at \(\mathbf{k}=\mathbf{0}\),
\(\mathbf{k}_{peak}\) is the location of the first dominant peak value of \(S\).
For more details, we refer to [HGBLachiezeR22] (Section 2.5).
Important
To compute \(\hat{S}(\mathbf{0})\), a linear extrapolation with a least-square fit is used to fit a line on the values of
sfassociated with a subvector ofk_norm. This subvector is obtained by truncatingk_normaroundk_norm_stop. For the choice ofk_norm_stop, the trade-off is to remain close to zero while including enough data points to fit a line. In addition,std_sfwill be considered while fitting the line if it’s not None.
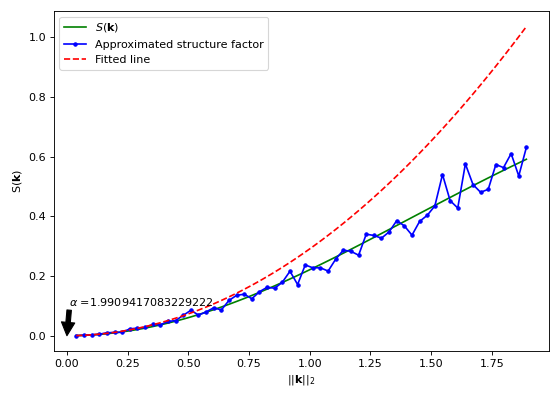
- structure_factor.hyperuniformity.hyperuniformity_class(k_norm, sf, k_norm_stop=1, std_sf=None, **kwargs)[source]
Fit a polynomial \(y = c \cdot x^{\alpha}\) to
sfaround zero. \(\alpha\) is used to specify the possible class of hyperuniformity of the associated point process (as described below).- Parameters
k_norm (numpy.ndarray) – Vector of wavenumbers (i.e. norms of the wavevectors).
sf (numpy.ndarray) – Evaluations of the structure factor, of the given point process, at
k_norm.std (numpy.ndarray, optional) – Standard deviations associated to
sf. Defaults to None.k_norm_stop (float, optional) – Threshold on
k_norm. The subvector obtained fromsfstarting from zero up to the value associated withk_norm_stopis used to fit a polynomial and find \(\alpha\). Defaults to 1.
- Keyword Arguments
kwargs (dict) –
Keyword arguments (except “sigma”) of scipy.scipy.optimize.curve_fit parameters.
- Returns
“alpha”: Estimated value of \(\alpha\).
”c”: Estimated value of \(c\).
”fitted_poly”: Polynomial used to find \(\alpha\).
- Return type
dict(float, float, function)
Example
import matplotlib.pyplot as plt from structure_factor.data import load_data from structure_factor.hyperuniformity import hyperuniformity_class from structure_factor.point_processes import GinibrePointProcess from structure_factor.structure_factor import StructureFactor from structure_factor.tapered_estimators_isotropic import ( allowed_k_norm_bartlett_isotropic, ) point_process = GinibrePointProcess() point_pattern = load_data.load_ginibre() sf = StructureFactor(point_pattern) d, r = point_pattern.dimension, point_pattern.window.radius k_norm = allowed_k_norm_bartlett_isotropic(dimension=d, radius=r, nb_values=60) k_norm, sf_estimated = sf.bartlett_isotropic_estimator(k_norm) summary = hyperuniformity_class(k_norm, sf_estimated, k_norm_stop=0.4) sf_fitted_0 = summary["fitted_poly"](k_norm) sf_theoretical = point_process.structure_factor(k_norm) fig, ax = plt.subplots(figsize=(7, 5)) ax.plot(k_norm, sf_theoretical, "g", label=r"$S(\mathbf{k})$") ax.plot(k_norm, sf_estimated, "b", marker=".", label="Approximated structure factor") ax.plot(k_norm, sf_fitted_0, "r--", label="Fitted line") ax.annotate( r"$\alpha$ ={}".format(summary["alpha"]), xy=(0, 0), xytext=(0.01, 0.1), arrowprops=dict(facecolor="black", shrink=0.0001), ) ax.legend() ax.set_xlabel(r"$||\mathbf{k}||_2$") ax.set_ylabel(r"$\mathsf{S}(\mathbf{k})$") plt.tight_layout(pad=1)
(Source code, png, hires.png, pdf)
 Definition
DefinitionFor a stationary hyperuniform point process \(\mathcal{X} \subset \mathbb{R}^d\), if \(\vert S(\mathbf{k})\vert\sim c \Vert \mathbf{k} \Vert_2^\alpha\) in the neighborhood of zero, then by [Cos21] (Section 4.1.) the value of \(\alpha\) determines the hyperuniformity class of \(\mathcal{X}\) as follows,
Class
\(\alpha\)
\(\mathbb{V}\text{ar}\left[\mathcal{X}(B(0, R)) \right]\)
I
\(> 1\)
\(\mathcal{O}(R^{d-1})\)
II
\(= 1\)
\(\mathcal{O}(R^{d-1}\log(R))\)
III
\(]0, 1[\)
\(\mathcal{O}(R^{d-\alpha})\)
For more details, we refer to [HGBLachiezeR22], (Section 2.4).
Important
To compute \(\alpha\), a polynomial is fitted on the values of
sfassociated with a subvector ofk_norm. This subvector is obtained by truncatingk_normaroundk_norm_stop. For the choice ofk_norm_stop, the trade-off is to remain close to zero while including enough data points to fit a polynomial. In addition,std_sfwill be considered while fitting the polynomial if it’s not None.
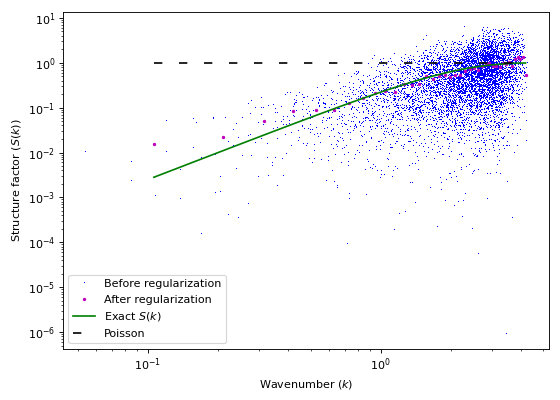
- structure_factor.hyperuniformity.bin_data(k_norm, sf, **params)[source]
Split
k_norminto sub-intervals (or bins) and evaluate, over each sub-interval, the mean and the standard deviation of the corresponding values insf.- Parameters
k_norm (numpy.ndarray) – Vector of wavenumbers (i.e. norms of wavevectors).
sf (numpy.ndarray) – Evaluations of the structure factor, of the given point process, at
k_norm.
- Keyword Arguments
params (dict) – Keyword arguments (except “x”, “values” and “statistic”) of scipy.stats.binned_statistic.
- Returns
k_norm: Centers of the bins.sf: Means of the structure factor over the bins.std_sf: Standard deviations of the structure factor over the bins.
- Return type
tuple(numpy.ndarray, numpy.ndarray, numpy.ndarray)
Example
import matplotlib.pyplot as plt import numpy as np import structure_factor.utils as utils from structure_factor.data import load_data from structure_factor.hyperuniformity import bin_data from structure_factor.point_processes import GinibrePointProcess from structure_factor.spatial_windows import BoxWindow from structure_factor.structure_factor import StructureFactor point_pattern = load_data.load_ginibre() point_process = GinibrePointProcess() # Restrict point_pattern to window window = BoxWindow([[-35, 35], [-35, 35]]) point_pattern_box = point_pattern.restrict_to_window(window) # Estimate S sf = StructureFactor(point_pattern_box) x = np.linspace(0, 3, 80) x = x[x != 0] k = utils.meshgrid_to_column_matrix(np.meshgrid(x, x)) k, sf_estimated = sf.scattering_intensity(k) # bin_data k_norm = utils.norm(k) k_norm_binned, sf_estimated_binned, _ = bin_data(k_norm, sf_estimated,bins=40) fig, ax = plt.subplots(figsize=(7, 5)) ax.plot(k_norm, sf_estimated, "b,", label="Before regularization", rasterized=True) sf.plot_isotropic_estimator( k_norm_binned, sf_estimated_binned, axis=ax, color="m", exact_sf=point_process.structure_factor, label="After regularization", ) ax.legend() plt.tight_layout(pad=1)
(Source code, png, hires.png, pdf)
Note
Typical usage
Regularize the results of
StructureFactorto be used ineffective_hyperuniformity()andhyperuniformity_class().
See also
- structure_factor.hyperuniformity.subwindows(window, subwindows_type='BoxWindow', param_0=None, param_max=None, params=None)[source]
Create a list of cubic (or ball)-shaped subwindows of a father window, with the associated minimum allowed wavevectors (or wavenumbers).
- Parameters
window (
AbstractSpatialWindow) – Father window.subwindows_type (str, optional) – Type of the subwindows to be created. The available types are “BoxWindow” and “BallWindow”. The former for cubic and the latter for ball-shaped subwindows. Defaults to “BoxWindow”.
param_0 (float, optional) – Parameter (lengthside/radius) of the first subwindow to be created. If not None, an increasing sequence of subwindows with parameters of unit increments is created. The biggest subwindow has parameter
param_maxif it’s not None, else, the maximum possible parameter. Defaults to None.param_max (float, optional) – Maximum subwindow parameter (lengthside/radius). Used when
param_0is not None. Defaults to None.params (list, optional) – List of parameters (lengthside/radius) of the output subwindows. For a list of parameters of unit increments,
param_0andparam_maxcan be used instead. Defaults to None.
- Returns
subwindows: Obtained subwindows.
k: Minimum allowed wavevectors of
allowed_k_scattering_intensity()or wavenumbers ofallowed_k_norm_bartlett_isotropic()associated with the subwindow list. The former is for cubic and the latter for ball-shaped subwindows.
- Return type
(list, list)
Example
from structure_factor.spatial_windows import BoxWindow from structure_factor.hyperuniformity import subwindows # Example 1: L = 30 window = BoxWindow([[-L / 2, L / 2]] * 2) # subwindows and k l_0 = 10 subwindows_box, k = subwindows(window, subwindows_type="BoxWindow", param_0=l_0) # Example 2: subwindows_params=[1, 4, 7, 15] subwindows_ball, k_norm = subwindows(window, subwindows_type="BallWindow", params=subwindows_params) import matplotlib.pyplot as plt fig, axis = plt.subplots(1, 2, figsize=(10,5)) axis[0].plot(0,0) axis[1].plot(0,0) for i, j in zip(subwindows_box, subwindows_ball): i.plot(axis=axis[0]) j.plot(axis=axis[1]) plt.show()
(Source code, png, hires.png, pdf)
Note
Typical usage
Create the list of subwindows with the associated k to be used in
multiscale_test().